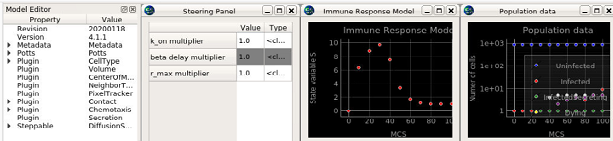
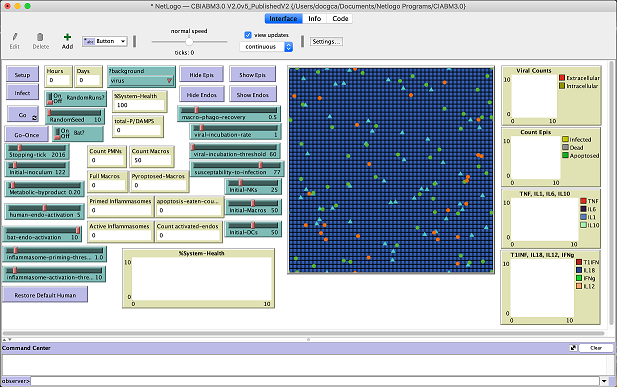
We have started a GLIMPRINT runnable models index page and individual pages for an intial set of three models.
One goal of GLIMPRINT is to create web-based resource where models relevant to immunology and digital twins can be hosted. Easy access to models in a runnable form is critical for education and for communicating the capabilities (and limitations) of computational models. The model should include a clear description of what is included, and should allow the user to run the model easily with the capability of modifying parameters and exploring outputs. We are particularly interested in the educational power of basic models that can be run interactively in a web browser.
Models will cover a range of complexities, from simple viral infection SIR (susceptible - infected - recovered) models in JavaScript or SBML running in a browser or Jupyter Notebook (or as a NetLogo model), to complex multiscale models that include discreet cells, subcellular processes, immune response, etc.
Our intial models are:
- A simple SIR model in JavaScript that runs directly in your browser.
- Sego et als. viral infection tissue (CC3D) model at nanoHUB.
- Cockrell and An's NetLogo model comparing infection and immune response in Humans and Bats (runs in browser or in a local NetLogo)
We hope to add many more models in the future. If you have a model you might like to share please let us know.