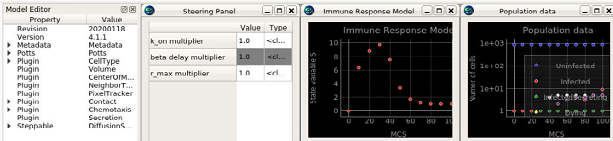
This model describes select interactions between generalized epithelial and immune cells and their extracellular environment associated with viral infection and immune response at the cellular and subcellular levels in a simulated epithelial layer.
How we built it: All model components are implemented in CompuCell3D, an open-source modeling and simulation environment of the spatiotemporal dynamics of virtual tissue at the cellular and subcellular levels. Model components under development are written in python and simulated in the CompuCell3D simulation engine, which is developed in C++. Supporting tools for documentation standards are being developed in python using Sphinx.
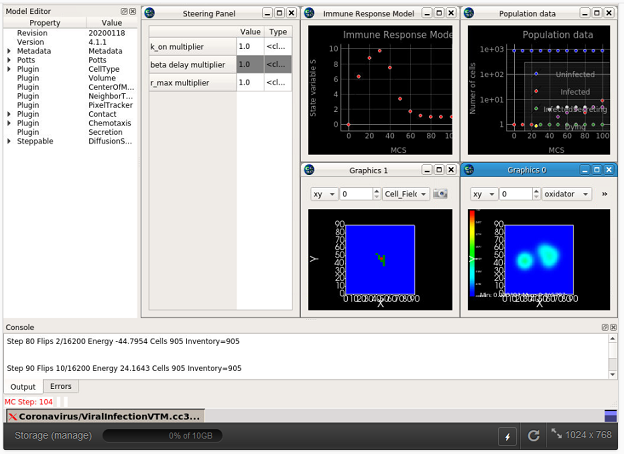
Model Location: This model can be run at the nanoHUB website (requires a nanoHUB account): https://nanohub.org/tools/cc3dcovid19. In addition, the entire project can be obtained from the github repository at https://github.com/covid-tissue-models/covid-tissue-response-models.