MSM Viral Pandemics meetings
April 18, 2023 Meeting
Zoom recording and slides are available for:
James Sluka, Indiana University, will discuss: "Predicting disease with 'broken' models, a work in progress."
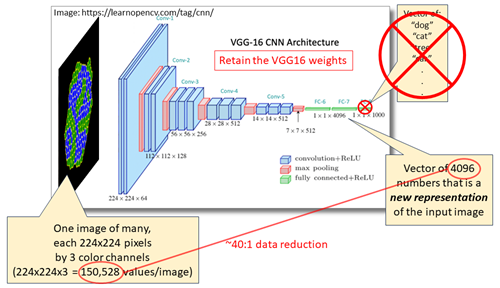
The development of computational models for biological processes often focuses on specific target states, such as normal and disease states. However, these models may have the capacity to represent additional states and outcomes. Indeed, the model may be able to represent multiple normal and disease states including those that the modeler did not develop their model for. Researchers often explore parameter space extensively, generating thousands of results. While numerical outputs can be clustered and classified using various approaches, simulations that primarily produce spatial models, such as 2D models, pose a different challenge. In such cases, results can be clustered based on user-defined metrics, like cell count or morphological characteristics. An alternative approach involves using modern image clustering tools based on artificial intelligence (AI) and machine learning (ML) algorithms. Here, I discuss the use of VGG16, a pretrained image classification AI/ML model, to recode simulation snapshots from a cell sorting Potts model. The recoded images, typically represented as short vectors, can be further simplified (e.g., using principal component analysis, PCA) and clustered (e.g., using hierarchical clustering) into sets of images. This approach provides an unbiased method for grouping a wide range of simulation results, especially when the simulation output primarily consists of images. We will compare this AI/ML-based clustering approach with clustering based on user-defined metrics for cell sorting.
YouTube and Slides (Note: The PowerPoint slides work best if you download a copy to your computer instead of viewing in a browser.)